ABSTRACT
Aiming at the difficulty of rice multi-scale pest and disease identification and deployment of lightweight detection models in field environment, this paper proposes a lightweight rice pest and disease identification model SCR-YOLO based on the improved YOLOv11n. The model carries out a triple optimization on the basis of YOLOv11n, the RepViT module is introduced to enhance the feature representation capability by structural reparameterization technique. The CBAM hybrid attention mechanism is embedded to strengthen the attention to the key areas of spots; and the SimSPPF module is adopted to optimize the efficiency of multi-scale feature fusion. CBAM hybrid attention mechanism to strengthen the focus on the key areas of disease spots, and adopting SimSPPF module to optimize the efficiency of multi-scale feature fusion. The experimental results on four types of rice pests and diseases datasets containing leaf blight, rice blast, hoary mottle and rice fly show that SCR-YOLO achieves a significant lightweight effect while maintaining a high detection precision, with a precision rate (P) of 84.7%, a recall rate (R) of 84.2%, a mean average precision of 87.9% (mAP50), a reduction in the number of model parameters to 2.3 M, and the computational effort is only 7.3 GFLOPs.Deployment tests on a Jetson Nano embedded device show that the improved model's single-image inference time is significantly optimized, achieving a detection speed of 8.3 frames per second. This study provides an efficient and feasible lightweight solution for real-time accurate identification of rice pests and diseases, which is of positive significance for promoting the practical application of intelligent plant protection equipment in complex field environments.
Keywords:Rice Pest Identification; Target Detection; YOLO11; Attention Mechanism; Lightweight Structure
INTRODUCTION
Rice is an important staple crop, and its stable yield growth is crucial for global food security. Among the various factors affecting rice production, pests and diseases are key limiting factors, and timely and accurate identification and control are essential to reducing losses. Currently, pest and disease monitoring and early warning systems for rice mainly rely on visual identification by farmers and laboratory testing. The former is inefficient and constrained by the distribution of human resources and professional levels [1], while the latter has high detection accuracy but is slow and expensive. Therefore, there is an urgent need to develop an efficient and intelligent automatic system for rice pest and disease identification. At present, crop pest and disease detection methods based on visual information processing mainly include image processing technology and deep learning techniques [2]. Image processing technology typically relies on manually extracting various image features such as color, texture, shape, and size [3-6]. However, these methods have limitations, such as being cumbersome to operate, low robustness, weak adaptability, poor accuracy, and time consumption. With the continuous improvement in computer performance and breakthroughs in deep learning technology, deep learning-based object classification and detection methods have been increasingly applied in agriculture [7-9]. The development of machine vision technology has also opened new avenues for crop disease detection. Pantazi et al. designed an automatic recognition system based on local binary patterns for identifying the health status of grape leaves and three diseases (downy mildew, powdery mildew, and black rot), achieving an overall classification success rate of 95%. Shin et al. attempted to combine three feature extraction methods—HOG, SURF, and GLCM—with ANN and SVM classifiers to identify strawberry powdery mildew, achieving the highest classification accuracy of 94.34% with ANN combined with SURF. Haruna et al. employed image analysis, sparse coding, artificial neural networks, and MATLAB tools for apple pest and disease classification, achieving an accuracy rate of 90%. These results show that traditional machine learning methods have good potential for disease recognition [10-12]. However, for crops like rice, due to the wide variety of pests and diseases and significant differences in disease spots, traditional image processing methods still face many difficulties in feature extraction, making it challenging to balance sensitivity in detecting small disease spots with classification accuracy for large-scale symptoms. In complex field environments, these methods are also easily affected by factors such as light changes and leaf occlusion, leading to unstable recognition performance. Additionally, most traditional models rely on manually designed features, limiting their generalization ability and making it difficult to meet the simultaneous classification needs of various rice diseases. In recent years, deep learning has provided new ideas for automatic pest and disease detection in rice. The team of Huang Shuangping applied Google Net for rice blast disease recognition, achieving an accuracy of 92.0%. Lin Xiangze et al. combined transfer learning with Mask R-CNN to propose a rice planthopper image classification scheme, achieving an average recognition accuracy of 92.3%. Bifta et al. used Faster R-CNN to identify rice leaf diseases, including rice blast, brown spot, and Hispa, with an accuracy of 99.25%. Zhang et al. developed a rice disease detection model based on ResNet, achieving a highest recognition accuracy of 99.87% for brown spot and rice blast diseases [13-16]. Yunusa et al. used the SSD algorithm based on data augmentation to achieve high-accuracy recognition of white leaf spot disease, rice blast, brown spot, and Tungro disease, with mean average precision (mAP) values of 0.91. Although the above studies perform well in feature extraction, models such as Faster R-CNN and SSD are generally large in terms of parameters and slow in inference speed, requiring high hardware resources. Therefore, how to achieve model lightweighting while maintaining recognition performance has become a key challenge in the current field of rice pest and disease detection [17]. To meet the deployment requirements of resource-constrained devices, researchers have carried out several lightweighting explorations. For example, Zhang et al. introduced a multi-scale attention mechanism based on the VGG16 model, improving the recognition accuracy to 92.44%. The YOLO algorithm proposed by Redmon et al. has significant advantages in terms of model parameters and detection speed [18,19]. Lü Shilei et al. proposed a lightweight network YOLOv3-LITE, using MobileNet-v2 as the backbone network, achieving nearly a 20-fold improvement in detection speed compared to Faster R-CNN. Chu Xin et al. proposed a MobileNetv1-YOLOv4 model based on YOLOv4, reducing model parameters by 80%. Zhou et al. combined Ghost Net with YOLOv4 for rice pest and disease recognition, reducing the model weight to 42.45MB. In addition, Jiao et al. proposed a YOLOv3-SPP model for lychee detection, achieving a compression rate of 96.8% through channel and layer pruning, with an average accuracy of 95.3%. Olarewaju et al. introduced ShuffleNetv2 into YOLOv5 for lightweighting, achieving mAP50 of 0.893 for fruit detection and segmentation tasks with only 3MB model weight. Indah et al. implemented lightweighting for YOLOv7 using Ghost Conv for logistics parcel detection, reducing model parameters to 30M and achieving an accuracy of 99.6%. However, lightweighting typically comes with a loss in accuracy. For example, Wang et al. proposed a lightweight module IPA for YOLOv5s, which significantly reduced the parameter size but caused a decrease in mAP50 by 0.01. Huang et al. combined MobileNetV3 and the SPPFCSPC-GS module for YOLOv7 lightweighting, resulting in a significant reduction in model parameters but a drop in mAP by 0.6%. Therefore, how to achieve model lightweighting while maintaining high accuracy and enabling edge deployment remains a significant challenge in current research. Based on the above background, this study selects rice white leaf spot disease, rice blast disease, rice brown spot, and rice planthopper as research objects, establishes a specialized image dataset, and proposes a recognition model SCR-YOLO that integrates multi-scale mechanisms [20,21]. This model optimizes the original network by introducing the RepViT structure, CBAM attention mechanism, and SimSPPF module in the backbone network. Further, through ablation experiments, performance comparison, and deployment testing, the effectiveness of the model improvements is validated, aiming to achieve precise recognition and real-time detection of multi-scale rice pest and disease identification under resource-limited hardware conditions.
MATERIALS AND METHODS
1.1 Dataset
The rice pest and disease identification dataset used in this study was constructed based on images captured in field environments. The dataset was collected from two major sources: the rice research station in Fang Zheng County, Harbin City, Heilongjiang Province, which provided RGB images of rice leaf diseases, and the experimental base at Northeast Agricultural University in Xiang fang District, Harbin City, which contributed images of rice planthoppers. The dataset contains images of four distinct types of rice leaf diseases: bacterial blight, rice blast, rice brown spot, and rice planthopper. In total, 6,739 rice leaf pest and disease images were collected in natural field conditions. However, due to challenges such as mixed diseases, blurriness, and other environmental interferences in the field, the images were filtered to eliminate poor-quality samples, resulting in a refined dataset of 6,588 images.
Figure 1: illustrates a selection of rice leaf disease images from the training set
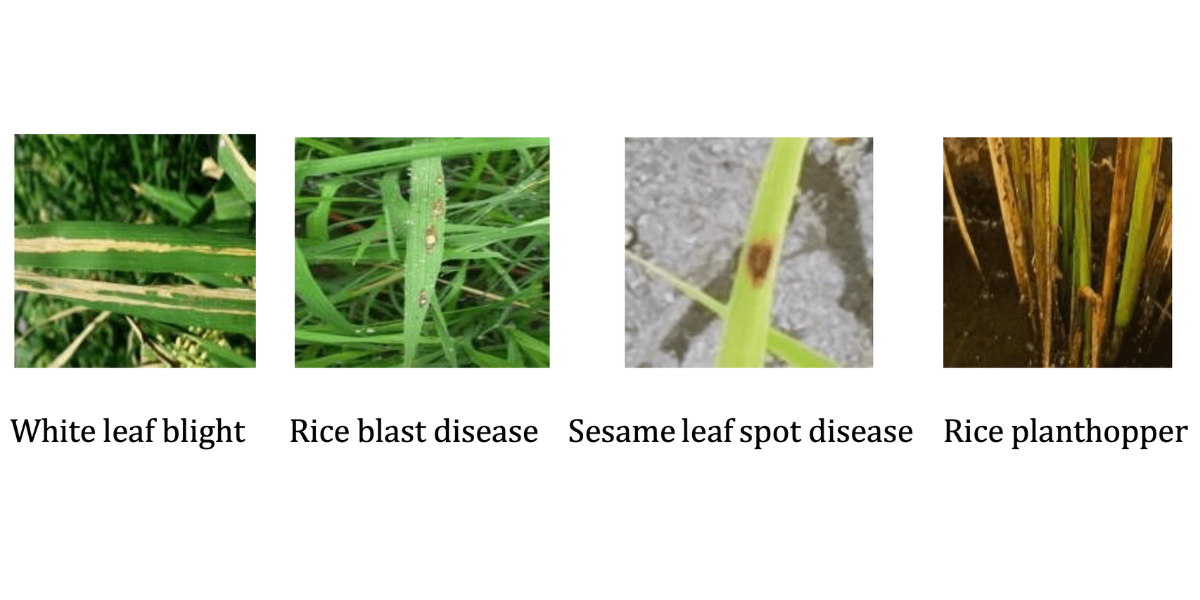
White leaf blight Rice blast disease Sesame leaf spot disease Rice planthopper
Figure 1: Images of rice leaf pests and diseases
1.2 Data Augmentation and Splitting
To further enhance the model’s generalization ability and recognition accuracy, several data augmentation techniques were applied to the retained images. These included adding noise, adjusting brightness, rotation, mirroring, and translation. After a second round of filtering, images with abnormal brightness, excessive blurring, or those that deviated significantly from realworld conditions were removed. As a result, a total of 13,176 high-quality images were selected for dataset construction, with examples of augmented images shown in Figure 2. The dataset was then split into train, Val, and test sets at a ratio of 7:2:1.
introduces several improvements aimed at enhancing both detection accuracy and computational efficiency. The key innovations are evident in the optimization of several critical modules. (1) Backbone Network, YOLO11 replaces the C2f module from YOLOv8 with a new C3K2 module. The C3K2 module is designed based on the CSP architecture and incorporates a more efficient bottleneck structure. By utilizing two consecutive 3×3 convolution operations in place of larger convolution kernels, this modification reduces computational complexity and parameter scale, while enhancing fine-grained feature extraction and small object modeling efficiency. (2) Addition of C2PSA Module: Following the SPPF module in the backbone network, YOLO11introduces the C2PSA module.
This module leverages a multi-head position-sensitive attention mechanism (PSA), incorporating spatial attention during the feature fusion stage. This enables the model to focus on key target regions in complex backgrounds, improving efficiency in scenarios like small object detection and background intricate patterns. (3) Detection Head Design: In the detection head, YOLO11 further optimizes the decoupled head with a lightweight design, adding two depthwise separable convolutions

Figure 2: Image after data enhancement
1.3 Method
1.3.1 YOLO11n Network Model
YOLO11, developed by Ultralytics, is a next-generation object detection model that builds upon the strengths of the YOLO series, offering high-efficiency real-time detection. YOLO11 (DWConv). This convolution operation significantly reduces both the model's parameter counts and computational load. While YOLO11 has demonstrated exceptional performance in general object detection tasks, its original backbone network without increasing computational complexity Additionally, we integrate the CBAM, which enables the model to dynamically adjust the attention mechanism across both channel and spatial dimensions. This helps suppress background noise from complex field conditions, enhancing the model's abilitó to focus on disease-affected areas. Furthermore, we replace the original SPPF (Spatial Pyramid Pooling Fusion) module with Simplified SPPF (SimSPPF). The simplified version reduces computational overhead while improving the fusion of multi-scale features. This modification strengthens the model's ability to detect small lesions and dense areas more accurately, particularly in scenarios with high visual complexity. The improvements introduced by RepViT, CBAM, and SimSPPF make the proposed model more effective in the precise detection of rice leaf diseases and pests in field environments, offering a robust solution for intelligent agriculture using deep learning techniues.
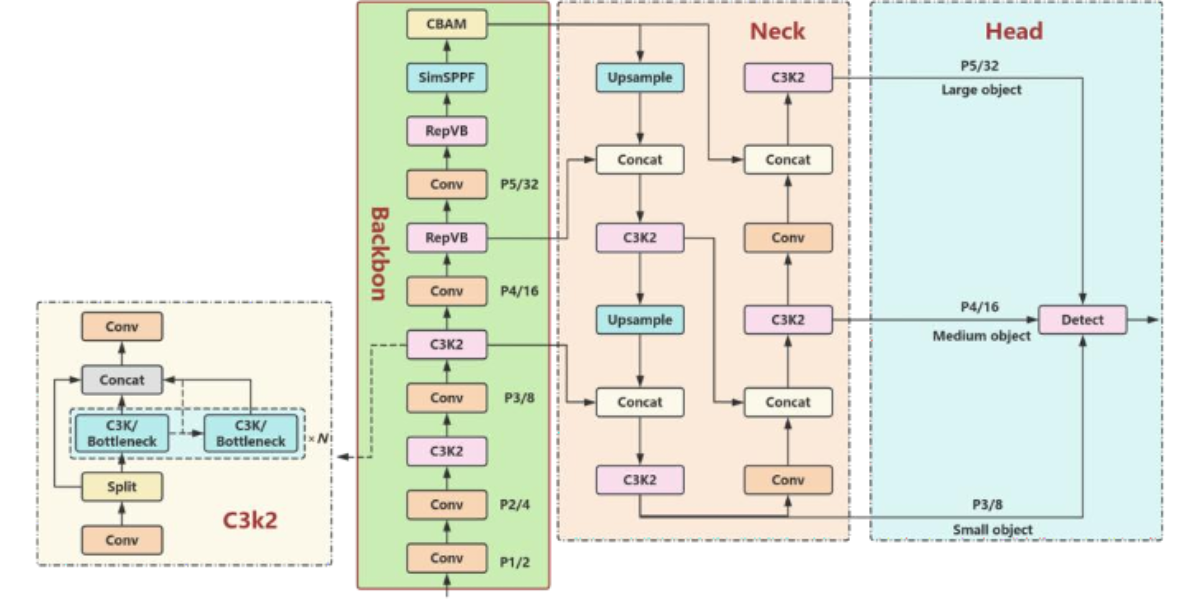
Figure 3: Improved YOLO11n model architecture diagram
1.3.3 Improvement of Feature Extraction Network
The introduction of the RepViT module into the YOLO11 backbone network for rice leaf disease and pest detection has significantly enhanced the model's performance, particularly in terms of detection accuracy and inference speed. RepViT is an innovative lightweight CNN architecture that integrates the efficient design principles of the Vision Transformer (ViT) into standards lightweight CNNs (such as MobileNetV3), progressively optimizing its adaptability for mobile device deployment. At the macro-architectural level, RepViT optimizes key components, such as the stem layer, down-sampling layers, classifiers, and overall stage proportions, to better cater to the requirements of mobile platforms. On the micro-architectural level, it fine-tunes aspects like convolution kernel size selection and the positioning of Squeeze-and-Excitation (SE) layers, ensuring more efficient use of computational resources and maintaining a lightweight structure. This targeted optimization allows the model to maintain high performance without compromising mobile device compatibility. From a modular perspective, RepViT employs structural reparameterization techniques that significantly enhance the learning efficiency during the training phase, while also reducing computational and memory overhead during inference. Additionally, the strategic placement of SE layers across blocks helps the model achieve optimal recognition accuracy while minimizing increases in latency. Various experiments have shown that RepViT outperforms existing lightweight ViT models across a range of vision tasks, demonstrating superior performance in both accuracy and inference latency. These findings underline the model's potential as a powerful tool for efficient disease and pest detection in precision agriculture, particularly in resourceconstrained environments.
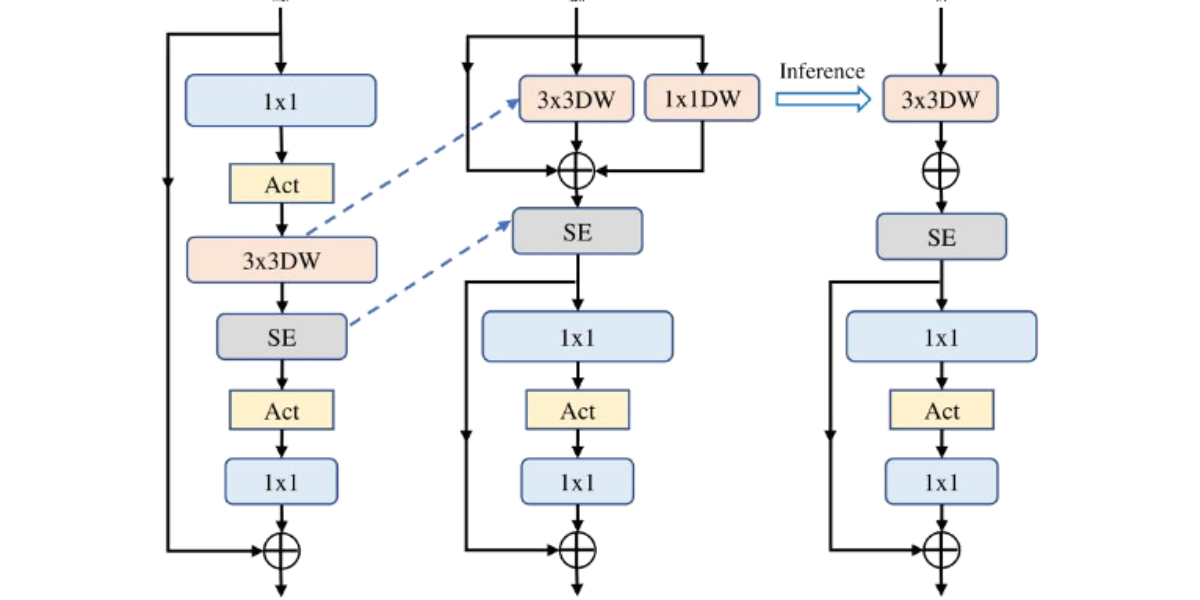
Figure 4: Schematic diagram of MobileNetV3 structure
Figure 4(a) illustrates the basic structure of MobileNetV3 with an optional Squeeze-and-Excitation (SE) layer. In Figure 4(b), through the application of structural reparameterization, the depthwise convolution and SE layer are rearranged, achieving the separation of the token mixer and the channel mixer. This method focuses on merging the original multi-branch structure into a single branch during the inference phase, thereby enhancing operational efficiency. The original MobileNetV3 block is composed sequentially of a 1×1 expansion convolution, depthwise convolution, and a 1×1 projection layer, with residual connections between the input and output. The SE module serves as an optional component, typically placed after the depth filter following the expansion convolution. Functionally, the 1×1 expansion convolution and projection layer are responsible for inter-channel information interaction, while the depthwise convolution focuses on spatial feature fusion. The former corresponds to the channel mixer, and the latter corresponds to the token mixer. In the original design, these two mixers are tightly coupled. To optimize the architecture, as shown in Figure 4(b), we move the depthwise convolution upward, effectively separating the token mixer from the channel mixer at the logical level. Additionally, during the training phase, structural reparameterization is introduced to enhance the representation power of the depth filter through a multi-branch topology. Since the SE module relies on spatial information, it is also moved to follow the depth filter to better utilize spatial contextual information in disease regions. Through this series of adjustments, the token mixer and channel mixer are effectively separated in the MobileNetV3 block. During inference, as shown in Figure 4(c), the multi-branch structure of the token mixer is consolidated into a single depthwise convolution operation. This significantly reduces the computational and memory overhead associated with skip.
connections and makes the model more suitable for deployment on mobile devices. The optimized module, referred to as the RepViT block, maintains its lightweight nature while significantly enhancing the semantic capture and spatiallocalization ability for multi-scale disease spots in rice leaf images. This improvement is particularly beneficial for increasing the detection accuracy and inference efficiency for disease areas, such as bacterial leaf blight and rice blast, in complex field backgrounds, offering effective support for mobile device deployment in smart agriculture applications.
1.3.4 Hybrid Attention Mechanism
The Convolutional Block Attention Module (CBAM) is a performance-enhancing module for convolutional neural networks (CNNs), consisting of two core sub-modules, the Channel Attention Module (CAM) and the Spatial Attention Module (SAM), as shown in Figure 7. These sub-modules operate by assigning attention weights to different channels and spatial positions, respectively, which enhances the model’s ability to represent features. Specifically, the CAM focuses on the feature information of the input image dataset, while the SAM attends to the spatial positional information of the image. CAM operates by compressing the spatial dimensions while preserving the channel dimensions, whereas SAM. compresses the channel dimensions while maintaining the spatial dimensions. The input features undergo parallel operations of max-pooling and average-pooling, followed by a compression-expansion transformation via a shared multi-layer perceptron in the channel attention module. The output of the channel attention is obtained through a sigmoid activation function and is multiplied by the input features to restore the original feature size. In the spatial attention module, the features obtained by multiplying the channel attention with the original image undergo both max-pooling and average-pooling operations. These pooled outputs are concatenated and passed through a 7×7 convolution to generate a single-channel feature map. Finally, the spatial attention output is derived using a sigmoid activation function and multiplied with the original image to restore the image to its original size.
In the equation, σ represents the sigmoid function, and x denotes the input features. The output Mc(F ) calculation formula of the channel attention module is as follows :
In the equation, F represents the input feature map, while AvgPool and MaxPool denote the global average pooling and max pooling operations, respectively. MLP represents the multilayer perceptron. The output M s (F ) calculation formula of the spatial attention module is as follows :
In the equation, f7×7 represents a 7×7 convolution operation, and [AvgPool(F); MaxPool(F)] concat denotes the concatenation of the results of the average pooling and max pooling along the channel axis.
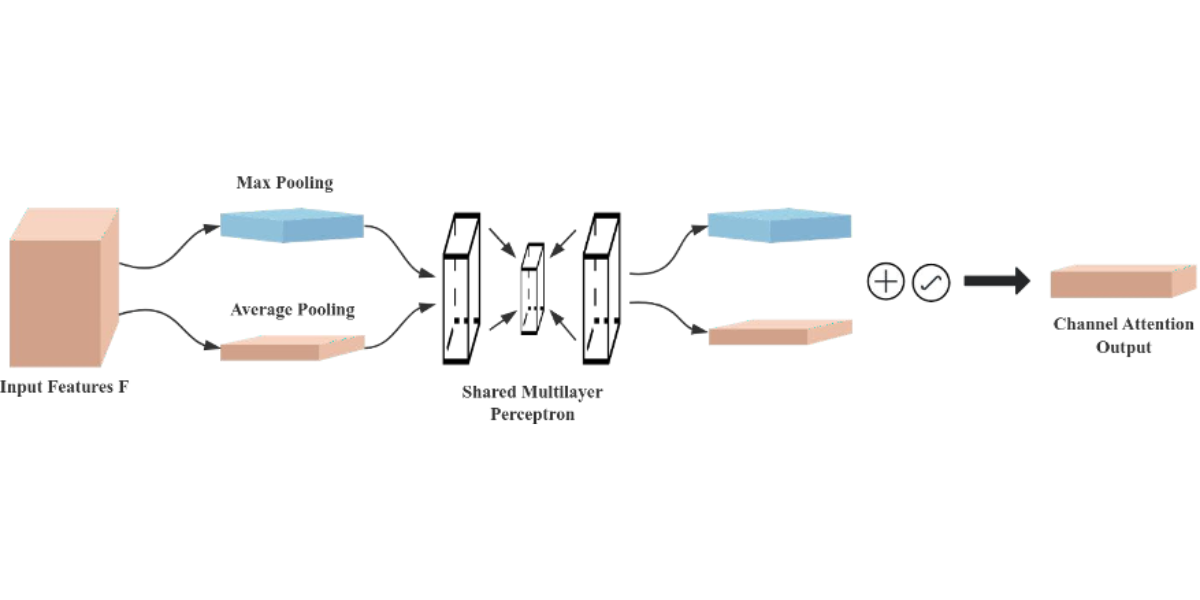
Figure 5: Schematic diagram of the channel attention module
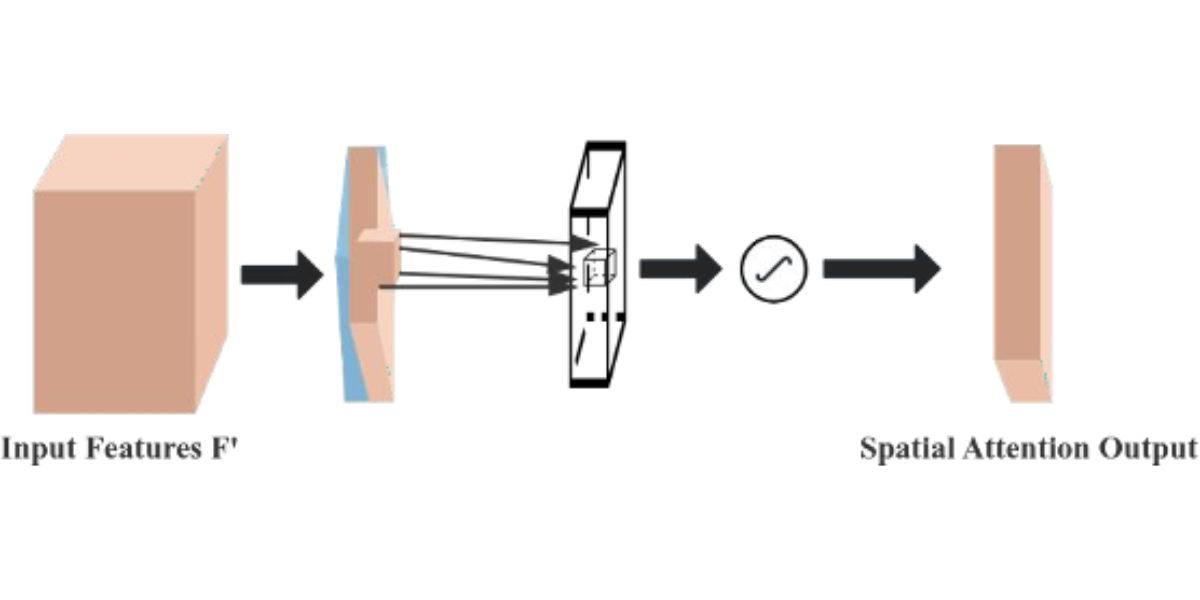
Figure 6: Schematic diagram of the spatial attention module
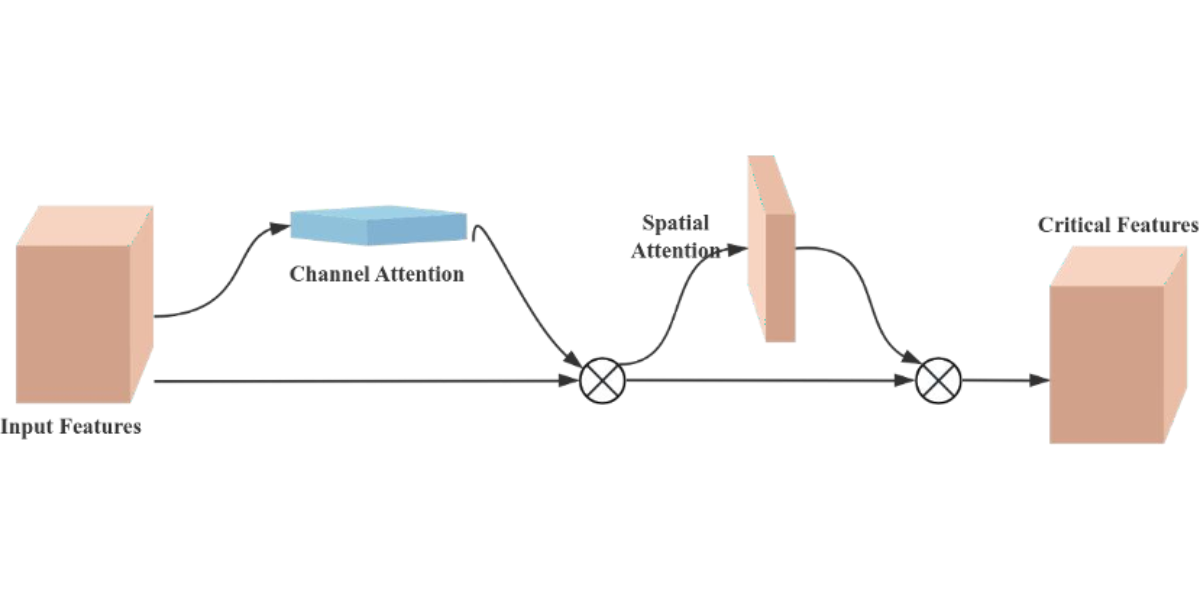
Figure 7: Schematic diagram of CBAM structure
1.3.5 Improvements in Feature Pyramid Networks
SimSPPF is a lightweight variant of SPPF (Spatial Pyramid Pooling Fast Layer), designed to enhance model efficiency by reducing redundant structures and parameter counts. This modification leads to significant improvements in computational performance while preserving feature extraction capabilities similar to those of the original SPPF. The architecture is illustrated in Figure 8. In the task of detecting rice leaf diseases and pests, SimSPPF efficiently handles regions with varying sizes of lesions, making it particularly well-suited for multi-scale target recognition in complex field environments. Unlike SPPF, which uses the SiLU activation function, SimSPPF adopts the ReLU function. This choice allows for further acceleration of computations while maintaining feature representation capabilities, thus providing effective support for the deployment of real-time disease detection systems in precision agriculture.
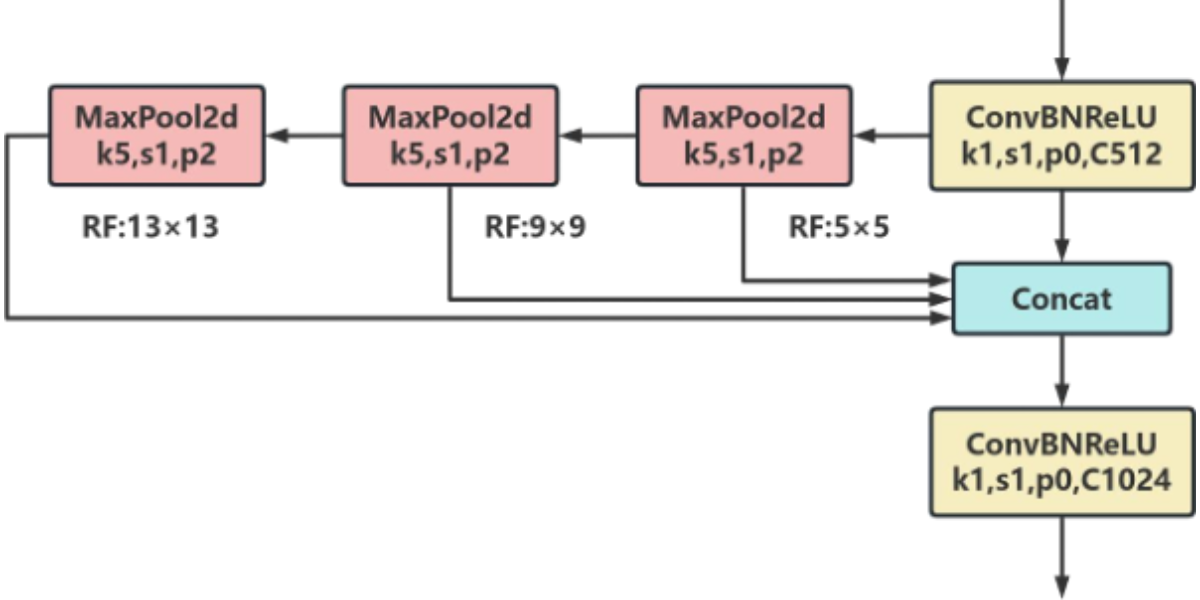
Figure 8: SimSPPF Network Architecture Diagram
2. Model assessme
2.0 Training Environment and Parameter Configuration
To comprehensively and objectively evaluate the performance of the proposed method, this study selected widely used and representative lightweight object detection models as comparison baselines, specifically YOLOv5n (a widely adopted classic lightweight model in the YOLO series) and YOLOv8n. All comparative and ablation experiments were conducted under a unified hardware and software environment, utilizing the PyTorch 2.4.1 deep learning framework, with training and testing performed on an NVIDIA GeForce RTX 4060 Laptop GPU. During the training process, identical dataset partitioning and hyperparameter settings were employed, including learning rate, batch size, and total number of epochs, to ensure fairnessin the comparison and reproducibility of results. The SGD optimizer was used, with momentum set to 0.937 and weight decay set to 0.0005. The number of epochs was set to 150, with a cosine annealing strategy applied for dynamic learning rate adjustment. The key hardware configuration is summarized in Table 1.
|
Environment Configuration |
Parameter GPU |
|
Operating System |
Windows |
|
CPU |
13th Gen Intel(R)Core(TM) i7-13650HX |
|
GPU |
NVIDIA GeForce RTX 4060 Laptop GPU |
|
GPU Memory |
8GB GDDR6 |
|
Operating Platform |
CUDA 12.6 |
|
Framework |
Pytorch 2.4.1 |
Table 1: Test environment configuration
2.1 Evaluation Metrics
To thoroughly assess the model's performance, this study developed a comprehensive evaluation framework that includes multiple dimensions such as detection accuracy, computational efficiency, and lightweight characteristics. The framework employs metrics like mean Average Precision (mAP@0.5:0.95, mAP@0.5, Precision, Recall, and F1-score to evaluate detection accuracy. Model complexity is measured using parameters and floating-point operations (FLOPs, while inference speed (FPS is recorded to reflect the model's deployment potential in realworld applications. To further analyze the individual and synergistic contributions of the RepViT module, CBAM module, and SimSPPF mechanism, systematic ablation studies were conducted. These experiments used the original YOLOv5n model as the baseline, sequentially introducing each module via a controlled variable approach. The experiments were performed on a standardized rice leaf disease dataset, with a quantitative evaluation of each module's effect. All ablation tests fixed hyperparameters such as random seed, input image size, batch size, initial learning rate, and training epochs to eliminate irrelevant interference. The final evaluation was based on a comprehensive assessment of the aforementioned accuracy and lightweight metrics.
Precision is calculated using the following formula:
Recall is calculated using the following formula:
The F1-score is calculated using the following formula:
The formulas for AP and mAP are as follows :
In the equation, AP@0.5 refers specifically to the AP value when the IOU threshold is 0.5, while AP@0.5:0.95 calculates the average AP by iterating the IOU threshold from 0.5 to 0.95 (with a step size of 0.05). This is used to evaluate the model's robustness under different localization precision requirements.
2.3 Model Results and Analysis
2.3.1 Comparative Experiments
In the target recognition and keypoint prediction models, the experiments were conducted using YOLOv5n, YOLOv8n, YOLO11n, and SCR-YOLO11n. The experimental results for each model are shown in Table 2. A comparison of the results reveals that the improved SCR-YOLO11n model outperforms the other baseline network models overall. The SCR-YOLO11n model achieves an mAP@0.50 of 87.9%, precision (P) of 84.7%, and recall (R) of 84.2%. Compared to the baseline model YOLO11n, the mAP@0.50 increases by 1.5 percentage points, precision improves by 1.0 percentage point, and recall increases by 4.5 percentage points. Additionally, SCR-YOLO11n has a parameter size of 2.3M and a computational load of 7.3G, maintaining its lightweight characteristics while achieving a comprehensive improvement in accuracy. The experimental results indicate that introducing the RepViT module into the backbone network to enhance feature extraction capabilities, incorporating the CBAM attention mechanism to focus on disease areas, and using the SimSPPF module to optimize multiscale feature fusion have effectively improved the model's accuracy and robustness in recognizing rice leaf diseases and pests. These improvements validate the effectiveness of the proposed approach.
|
Target Recognition Performance |
Model Efficiency |
||||||
|
Model |
P (%) |
R (%) |
F1-score (%) |
mAP @0.50 (%) |
mAP@ 0.50:0.95 (%) |
Parameters (M) |
FLOPs (G) |
|
SCR-YOLO11n |
84.7 |
84.2 |
83 |
87.9 |
53.9 |
2.3 |
7.3 |
|
YOLOv8n |
82.8 |
79.2 |
82 |
86.2 |
51 |
3.2 |
8.9 |
|
YOLOv5n |
82.1 |
77 |
79 |
84.3 |
47.2 |
2.7 |
7.8 |
|
YOLO11n |
83.7 |
79.7 |
82 |
86.4 |
51.2 |
2.6 |
6.6 |
Table 2: Comparative test
2.3.2 Ablation Study on the Improved YOLO11n
In the same experimental setup, the SCR-YOLO11n model, which incorporates the RepViT, CBAM, and SimSPPF modules, achieved the best overall performance. Specifically, its precision (P) was 84.7%, recall (R) was 84.2%, F1-score was 83.0%, mAP@0.50 reached 87.9%, and mAP@0.50:0.95 was 53.9%. Compared to the baseline YOLO11n, this combination significantly reduced the model's parameter count while improving detection accuracy across all metrics. This suggests that the modules function effectively in a complementary and synergistic manner. The detailed experimental results are presented in Table 3.
The RepViT module, as a core component of the backbone network, is primarily responsible for enhancing global feature extraction capabilities. When added alone, the parameter count decreased from 2.6M to 2.5M, recall improved from 79.7% to 82.1%, and mAP@0.50:0.95 increased from 51.2% to 52.7%. This indicates that the RepViT module, through a reparameterization design, effectively broadened the model's ability to perceive disease regions. However, this improvement came at the cost of an increase in computational complexity, with FLOPs rising from 6.6G to 7.5G. The CBAM attention mechanism focuses on optimizing feature selection efficiency. When used alone, it had the most significant impact on model lightweighting, reducing parameters to 2.4M and FLOPs to 6.3G. Although precision slightly decreased, recall increased to 81.0%, indicating that the dual-attention mechanism of CBAM efficiently focused on critical disease regions while improving detection sensitivity without heavily taxing computational resources. The SimSPPF module primarily enhances multi-scale feature fusion capabilities. When used alone, it led to notable improvements in precision (84.3%), recall (82.8%), and mAP@0.50:0.95 (53.8%), with only a slight increase in computational cost (FLOPs 6.4G). This demonstrates that the SimSPPF module effectively enhanced the model's ability to identify lesions of varying sizes by optimizing the feature pyramid structure. When all three modules were combined, the SCR-YOLO11n model reached optimal performance. RepViT provided a strong foundation for feature extraction, CBAM optimized feature selection efficiency, and SimSPPF enhanced multi-scale perception, forming a complete technical loop. The final model, maintaining its lightweight characteristics parameter count of 2.3M), achieved the best results across all accuracy metrics.The systematic ablation study results show that the coordinated use of RepViT, CBAM, and SimSPPF modules significantly enhanced the performance of the YOLO11n model for rice leaf disease detection. The final model strikes an ideal balance between accuracy and efficiency, providing an effective technical solution for real-time crop disease identification in complex field environments. These results further validate the feasibility and practicality of the proposed algorithm improvements.
As shown in the comparison of detection results in Figure 9, the improved SCR-YOLO11n model demonstrated significant enhancements in visual perception compared to the original YOLO11n model. In handling rice pests and diseases with diverse morphologies and significant scale differences, SCRYOLO11n exhibited clear advantages. In the detection of bacterial blight, the global representation capabilities of the RepViT module, in collaboration with the CBAM attention mechanism, greatly improved the accuracy of detecting leaf edge yellowing and striped disease symptoms, effectively distinguishing the disease-healthy boundary areas. For the common multi-scale lesions in rice blast, the SimSPPF module, through its optimized multi-scale feature fusion mechanism, significantly improved continuous detection of both small spots and large diseased areas, while preserving the integrity of feature extraction. In the recognition of rice grain smut, the CBAM attention mechanism effectively suppressed background interference from leaf texture, allowing the model to focus precisely on the dense distribution of small disease spots, significantly reducing the false negative rate for small lesions. For small target pests like the rice planthopper, the enhanced feature extraction capability of RepViT and the multiscale perception properties of SimSPPF complemented each other, ensuring stable detection even in challenging scenarios where the pests' color closely matched the leaves. These visual improvements were consistent with the quantitative performance metrics, demonstrating that the SCR-YOLO11n model not only performed excellently in numerical evaluations but also exhibited enhanced environmental adaptability and robustness in real-world detection tasks. This makes it a reliable technical solution for precise identification of rice diseases and pests in complex field environments.
|
Target Recognition Performance |
Model Efficiency |
|||||||||
|
Baseline Model |
RepViT |
CBAM |
SimSPPF |
P (%) |
R (%) |
F1-score (%) |
Map (%) |
mAP@ (%) |
Parameters (M) |
FLOPs (G) |
|
|
× |
× |
× |
83.7 |
79.7 |
82 |
86.4 |
51.2 |
2.6 |
6.6 |
|
|
√ |
× |
× |
83.5 |
82.1 |
83 |
87.2 |
52.7 |
2.5 |
7.5 |
|
|
× |
√ |
× |
82.6 |
81 |
82 |
86.8 |
51.5 |
2.4 |
6.3 |
|
YOLO |
× |
× |
√ |
84.3 |
82.8 |
83 |
87.6 |
53.8 |
2.6 |
6.4 |
|
11n |
√ |
√ |
× |
82.8 |
83.5 |
83 |
87 |
52.3 |
2.3 |
7.3 |
|
|
√ |
× |
√ |
83.6 |
84 |
83 |
87.4 |
53.7 |
2.5 |
7.5 |
|
|
× |
√ |
√ |
84.5 |
82.3 |
83 |
87.6 |
53.6 |
2.3 |
6.3 |
|
|
√ |
√ |
√ |
84.7 |
84.2 |
83 |
87.9 |
53.9 |
2.3 |
7.3 |
Table 3: Ablation test
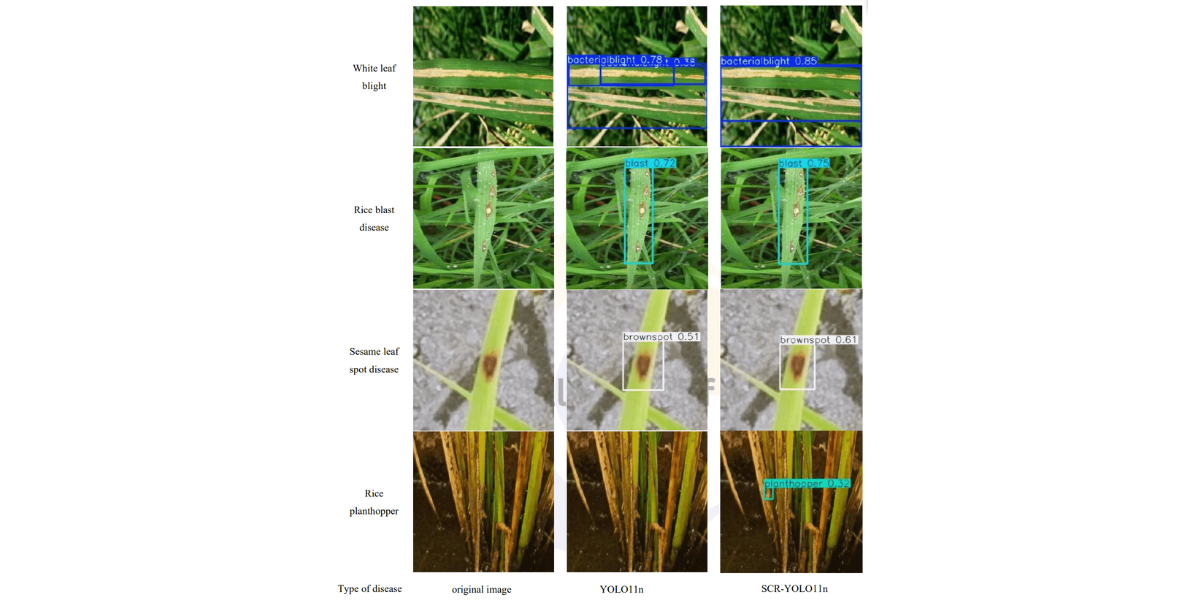
Type of disease original image YOLO11n SCR-YOLO11n
Figure 9: Comparison of model detection effect before and after improvement
2.3.2 Model Feature Visualization
To further investigate the decision-making process of the model in identifying rice leaf diseases and pests, this study utilizes the GradCAMPlusPlus method to generate class activation heatmaps, as shown in Figure 10. This visualization technique provides an in representation of the model's focus on different regions of the image during target detection by analyzing the variations in color intensity on the heatmap, we can clearly observe how the network responds to different areas of the image. The experimental results show that the original YOLO11n model tends to produce false negatives in complex scenarios, such as when disease spots closely resemble the background color or when there is leaf occlusion. In these cases, the heatmap response regions are often scattered, and the model pays insufficient attention to small lesions and areas with blurred edges. In contrast, the improved SCR-YOLO11n model exhibits a more precise attention distribution. It not only focuses more accurately on small, edge-blurred lesion regions but also demonstrates enhanced capability distinguishing densely distributed or partially overlapping disease spots. From the heatmap comparisons, it is evident that the SCR-YOLO11n model shows more concentrated and relevant activation responses in critical regions of various diseases, wherea the original YOLO11n model's attention points are relatively dispersed and less precise. This visualization confirms that by incorporating the RepViT, CBAM, and SimSPPF modules, the SCR-YOLO11n model significantly improves its ability to extract disease-related features and enhances spatial localization accuracy. As a result, the model demonstrates more reliable reliable identification performance and decision interpretability in complex field environments.
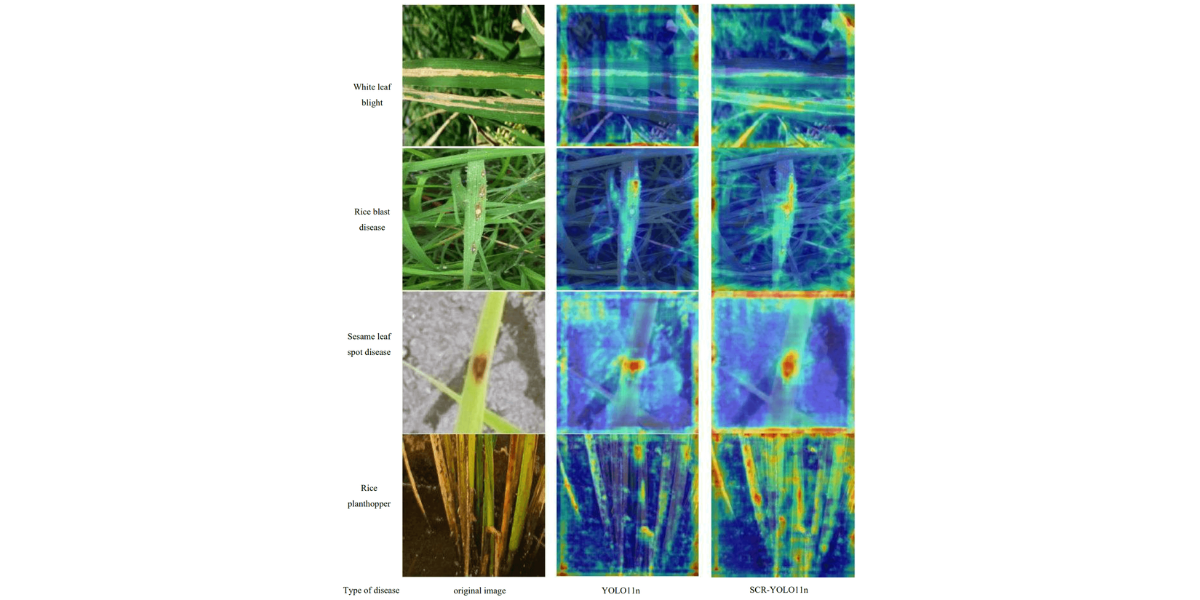
Figure 10: Heat map of model detection effect before and after improvement
DISCUSSION
This study successfully developed and validated the SCRYOLO11n, a rice leaf disease and pest detection model based on an improved version of YOLO11n. Ablation test results indicate that the integrated modules complement each other effectively, enhancin the overall performance of the model. The final model achieved an mAP@0.50 of 87.9%, recall of 84.2%, and precision of 84.7%, significantly outperforming the baseline YOLO11n model. Furthermore, it outperforms other mainstream lightweight detection architectures, such as YOLOv5n and YOLOv8n, in similar agricultural disease detection tasks. The RepViT module, through reparameterization, strengthened the model's global feature extraction capability, while the CBAM attention mechanism improved the model's focus on critical disease regions. Additionally, the SimSPPF module optimized multi-scale feature fusion efficiency, and the collaboration of these three components markedly enhanced the model's robustness in detecting small lesions, overlapping symptoms, and other complex scenarios. While improvingaccurac, the SCR-YOLO11n maintains a relatively low parameter count and computational cost, with 2.3M parameters and 7.3 GFLOPs of computational load. When compared to models such as Zhang et al.'s [18] VGG16-based improvement, Zhou et al.'s YOLOv4- GhostNet method, and Olarewaju et al.'s ShuffleNetv2- based lightweight solution this model demonstrates a clear advantage in terms of accuracy-efficiency trade-off. Notably, when compared with pruning methods like those of Jiao et al. and GhostConv-based modifications by Indah et al. this model achieves significant weight reduction without sacrificing accuracy, striking a balance between "high accuracy" and "lightweight" deployment. This makes it highly feasible for field deployment in agricultural settings. Compared to existing research, the innovation of this work lies in its approach, which goes beyond the direct application of general detection models. Instead, it addresses the unique challenges posed by the rice leaf disease detection task, such as variable scale and complex background, by systematically optimizing the model architecture. Unlike previous works by Lv Shilei et al. and Chu Xin et al. [21], which focused on lightweighting breplacing backbone networks, or those relying on post-processing compression techniques like pruning and quantization, this study introduces efficient components such as the RepViT reparameterization module and CBAM attention mechanism during the design phase. This strategy optimizes both accuracy and efficiency from the outset, providing new insights into the design of lightweight detection models for agricultural applications. Future research will focus on evaluating the generalization ability of the model across different rice cultivation regions and growth stages. The exploration of automated model optimization using neural architecture search (NAS) and the incorporation of multispectral and other multimodal data will be key to further enhancing the model's robustness in complex environments. Ultimately, this will facilitate the model's practical application in agricultural drones, field inspection robots, and other edge devices, providing reliable technical support for the advancement of intelligent crop protection systems.
CONCLUSION
This study introduces an enhanced YOLO11n model, SCRYOLO11n, which integrates the RepViT, CBAM, and SimSPPF modules for rice leaf disease and pest detection in complex field environments. Ablation test results demonstrate that the proposed model outperforms the baseline model across several key metrics. Specifically, precision (P) improved from 83.7% to 84.7%, recall (R) increased from 79.7% to 84.2%, and the mAP@0.50 reached 87.9%, a 1.5 percentage point improvement over the baseline model (86.4%). Furthermore, mAP@0.50:0.95 rose from 51.2% to 53.9%. The model not only maintains lightweight characteristics but also achieves excellent detection performance, with a parameter count of 2.3M and computational load of 7.3 GFLOPs, making it suitable for edge deployment in agricultural applications. Feature response and visualization analysis further confirm that the integrated modules effectively enhance the model's ability to perceive and discriminate multi-scale lesions. This enables the model to handle complex field conditions, such as leaf occlusion, lighting variations, and overlapping lesions, more effectively.
CRediT authorship contribution statement
Huijie Li: Conceptualization; Data curation; Formal analysis;Investigation; Methodology; Resources; Software; Supervision; Validation; Visualization; Writing-original draft; Writing-review & editing.
Li Xiao: Conceptualization; Formal analysis; Resources; Supervision; Visualization; Writing-review & editing.
Yanling Yin: Data curation; Formal analysis; Supervision.
Jingbin Li: Data curation; Investigation; Validation. Huanhuan Wang: Data curation; Software; Visualization.
Yang Li: Data curation; Supervision.
Hongfei Yang: Formal analysis; Investigation; Methodology; Software.
Zhentao Wang: Conceptualizatio Formal analysis; Funding acquisition; Project administration; Resources; Supervision; Visualization; Writing-review & editing.
ACKNOWLEDGMENTS
This work was conducted with the support of the High-level Talents Research Initiation Project of Shihezi University (RCZK202559), Project of Tianchi Talented Young Doctor (CZ002559), Tianchi Talent Program of Xinjiang Uygur Autonomous Region (2025, CZ000208) and the Shihezi University High-Level Talents Research Start-up Fund Program (RCZK2025105), Key Areas Science and Technology Research Project of Xinjiang Second Division (2025GG2601,2025GG2702) and China's National Key R &D Plan (2021YFD200060502).
Data Availability
Data will be made available on request
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- Liao J, Tao W Y, Zang Y, et al. (2023). Research progress and prospect of key technologies in crop disease and insect pest monitoring. Tansactions of the Chinese Society for Agricultural Machinery, 54, (11): 1-19. [Google Scholar]
- Ngugi L C, Abelwwahab M, Aao--Zahhad M. (2021). Recent advances in image processing techniques for automated leaf pest and disease recognition-a review. Information Processing in Agriculture, 8(1): 27-51. [Google Scholar]
- Padol P B,Sawant S D. (2016). Fusion classification technique used to detect downy and powdery mildew grape leaf diseases. International Conference on Global Trends in Signal Processing, Information Computing and Communication. Jalgaon: IEEE, 2016. [Google Scholar]
- Dey A K, Sharma M, Meshram M R. (2016). Image processing-based leaf rot disease, detection of betel vine (Piper betle L). Procedia Computer Science, 85: 748-754. [Crossref] [Google Scholar]
- Tian Y N, Yang G D, Wang Z, et al. (2020). Instance segmentation of apple flowers using the improved mask R-CNN model. Biosystems Engineering, 193: 264-278. [Crossref] [Google Scholar]
- Wang X W, Zhao Q Z, Jiang P, et al. (2022). LDS-YOLO: A lightweight small object detection method for dead trees from shelter forest. Computers and Electronics in Agriculture, 198: 107035. [Crossref] [Google Scholar]
- Pantazi X E, Moshou D, Tamouridou A A. (2019). Automated leaf disease detection in different crop species through image features analysis and one class classifiers. Computers and Electronics in Agriculture, 156: 96-104. [Crossref] [Google Scholar]
- Jaemyung Shin,Young K. Chang,Brandon Heung, et al. (2020). Effect of directional augmentation using supervised machine learning technologies: A case study of strawberry powdery mildew detection. Biosystems Engineering, 194: 49-60. [Crossref] [Google Scholar]
- Yousef A G, Abdollah A, Mahdi D, et al. (2022). Feasibility of using computer vision and artificial intelligence techniques in detection of some apple pests and diseases. Applied Science, 12, (2): 906-906. [Crossref] [Google Scholar]
- Huang S P, Sun C, Qi L, et al. (2017). Rice panicle blast identification method based on deep convolution neural network. Transactions of the Chinese Society of Agricultural Engineering, 33(20): 169-176. [Google Scholar]
- Bari B S, Islam M N, Rashid M, et al. (2021). A real-time approach of diagnosing rice leaf disease using deep learning-based faster R-CNN framework. Peerj Computer Science, 7, e432. [Crossref] [Google Scholar] [PubMed]
- Zhang Y J, Zhong L T, Ding Y, et al. (2023). ResViT-Rice: A deep learning model combining residual module and transformer encoder for accurate detection of rice diseases. Agriculture, 13(6): 1264-1264. [Crossref] [Google Scholar]
- Haruna Y, Qin S Y, Kiki M J, et al. (2023). An improved approach to detection of rice leaf disease with GAN-Based data augmentation pipeline. Applied Sciences, 13(3): 1346-1346. [Crossref] [Google Scholar]
- Redmon J, Divval S, Girshick R, et al. (2016). You only look once: Unified, real-time object detection In: IEEE/CVF 2016 IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, NV: IEEE, 2016. [Crossref] [Google Scholar] [PubMed]
- Zhou W, Niu Y Z, Wang Y W, et al. (2022). Rice disease and pest recognition method based on improved YOLOv4-GhostNet. Journal of Jiangsu Agricultural Sciences, 38(3): 685-695. [Google Scholar]
- Jiao Z Y, Huang K, Jia G Z, et al. (2022). An effective litchi detection method based on edge devices in a complex scene. Biosystems Engineering, 222: 15-28. [Crossref] [Google Scholar]
- Lawal O M. (2023). YOLOv5-LiNet: a lightweight network for fruits instance segmentation. Plos One, 18: e0282297. [Crossref] [Google Scholar] [PubMed]
- Firdiantik A I M, Lee S, Bhattacharyya C, et al. (2024). EGCY-Net: an ELAN and GhostConv-Based YOLO network for stacked packages in logistic systems. Applied Sciences, 14: 2763-2763. [Crossref] [Google Scholar]
- Wang Y, Xu S, Wang P, et al. (2024). Lightweight Vehicle Detection Based on Improved YOLOv5s. Sensors, 24, 1182. [Crossref] [Google Scholar] [PubMed]
- Huang D, Tu Y, Zhang Z, et al. (2024). A lightweight vehicle detection method fusing GSConv and Coordinate attention mechanism. Sensors, 24, 2394. [Crossref] [Google Scholar] [PubMed]
- Zhao H S, Zhang Y, Liu S, et al. (2018). PSANet: point-wise spatial attention network for scene parsing. Proceedings of the 15th European Conference on Computer Vision. Munich: Springer, 2018: 270-286. [Google Scholar]