Indexed In
Journal Flyer

Journal Highlights
Useful Links
Recommended Journals
Open Access Journals
ABSTRACT
Early and accurate detection of pests and pathogens is crucial factor to restrict the spread of rising infections that could otherwise cause remarkable economic losses. One such approach is POC diagnostics, that are fast, accurate, affordable, and reliable technologies performed on site even by a minimally trained-personnel, using minimal equipment. Among the available diagnostic technologies, Nucleic acid-based methods have emerged as a good choice for the development of detection tools in several fields, such as human/animal health, food safety, and plant pathogen detection. However, following PCR based assays, multiple types of nucleic acid amplification technologies have been developed such as loop-mediated isothermal amplification, helicase-dependent amplification, rolling circle amplification, recombinase polymerase amplification, that eliminated the requirement for thermocycling, making them more suitable for use as POC. These amplification methods along with POC sequencing have revolutionized diagnostics with their exceptional specificity and sensitivity, making them precious tool for precise diagnosis. The aim of this POC diagnostics is to ensure molecular detection of plant pathogens accessible to anyone, anywhere, and at any time. The success and usefulness of POC amplification are ultimately dependent on the availability of POC-friendly nucleic acid extraction methods and amplification readouts. Keywords: Isothermal amplification; point-of-care diagnostics; pathogen; sensitivity; specificity
INTRODUCTION
Plant pathogens pose a significant threat to agriculture worldwide, resulting in substantial reduction of crop productivity. According to the Food and Agriculture Organization (FAO), 20-40 percent of global crop production is lost to diseases, that costs approximately USD $220 billion every year (FAO,2021). This emphasizes the dire need to prioritize disease management in agriculturally based economies and increase global crop production by 20- 40% by just defeating pests and diseases. In the absence of resistance, the best alternative is to detect pathogens in the field early to prevent the onset of the disease [1]. The conventional approach of detecting and identifying plant pathogens are based on Koch’s postulates that requires examination of symptoms and culture-based methods. These methods usually take days or even weeks. Furthermore, identification by studying morphological characters of pathogen via microscopy demands specifically experienced pathologist. Although conventional approach is reliable, but at the same time it is lengthy and time-consuming process. Consequently, there is a dire need to develop new detection tools offering high sensitivity, specificity and speedy identification of plant pathogens. Since the 1980s, antibodybased approaches have greatly advanced on-site detection and diagnostics, becoming a prominent and robust tool due to their speed, sensitivity and cost effectiveness [2]. The techniques like immunoblots and enzyme-linked immunosorbent assays (ELISA) are indeed precise and sensitive, but not suitable for field applications. The Antibody-based lateral flow devices (LFDs) are quite suitable for point-of-care (POC) use, but are less sensitive compared to nucleic acid–based techniques. During 1980s, the birth of PCR restructured diagnostics, and increasing number of laboratories embraced DNA based techniques for highly specific and extremely sensitive detection and identification of pathogen-specific DNA sequences. As a result, a plethora of PCR-based detection protocols have been documented for the identification of plant pathogens. Furthermore, new approach to enhance specificity and sensitivity of pathogen detection involves coupling of PCRS with other techniques. For example, early 1990s saw the development of PCR-ELISA, a combination of conventional PCR and enzyme-linked immunosorbent assay (ELISA), that was effectively utilized to detect viruses, bacteria and fungi with higher specificity in comparison to conventional PCR. An introduction of real-time quantitative PCR was the major advancement in DNA-based detection techniques, enabling both detection as well as quantification of pathogen levels in samples, hence permitting the assessment of pathogen infection severity. while these PCR based methods are reliable, specific, robust and have been successfully applied for detection of multiple pathogens at the same time, but they still possess drawbacks like need of costly equipment’s and reagents as well as trained personnel. Moreover, during multiplex detection of unknown pathogens, these techniques are still prone to nonspecific DNA amplification, fetching false positive results hence, limiting its suitability for POC application. Nucleic acid-based detection has undergone a revolution with the introduction of next-generation sequencing [3]. The use of bioinformatics tools helps in primer designing for detection of single pathogen, mutated variant and multiple pathogens in a single assay [4].
POC detection of plant pathogens
The rapid pathogen detection techniques are creating possibilities for development of novel tools, which can carry straight to the intended analysis site, referred as point of-care (POC). So, there is growing demand for POC tests that do not need complicated instruments, allowing rapid and cheap testing in the field [6]. POC tools should consist of reliable, portable, easy to use, and affordable equipment’s, having capacity of conducting all the required steps swiftly and with minimal complexity, even performed by those with only little training [7]. The World Health Organization (WHO) has consolidated these requirements into the ASSURED (Affordable, Sensitive, Specific, User-friendly, Robust and rapid, Equipment-free, Deliverable) guidelines for POC testing. Agricultural industries gain a great deal by using affordable POC tests as crop field are located far from laboratory setups and sample transport might present logistical challenges. Despite integrating every step of the process, from sample preparation to result visualization, a POC diagnostic technology still presents difficulties [8]. The most promising options for POC analysis protocol are DNA-based techniques. It mainly comprises of three key steps: (a) on-site nucleic acid extraction and purification (b) nucleic acid amplification (c) result visualization. The effectiveness of amplification technique is closely linked to the initial extraction step, as low nucleic acid yield or presence of inhibitors can sometimes provide false-negative results. Similarly, result visualization is also innately associated to the amplification step,as some techniques need comparatively simple visualization, whereas others require complex instruments [9]. In this review, we researched the major breakthrough in DNA-based technologies for developing portable instruments tailored for detection and identification of plant pathogens, with an emphasis on their use in agriculture [10].
Advantages of POC testing
• Facilitate thorough and rapid plant screening to avoid spread of disease [11].
• On site analysis of imported plants and products before they can enter a country to prevent disease introduction.
• Investigation and researching newly introduced non-native pathogens.
• Disease surveillance [12].
POC techniques for nucleic acid extraction
The adoption of DNA amplification technologies for POC analysis, one of the major obstacles is the extraction of nucleic acid [13]. The high-quality DNA or RNA is always important to acquire reliable and repeatable results, regardless of any nucleic acid amplification method used. The aim of this process is to extract as much of the nucleic acid as possible from the pathogen while simultaneously minimizing impurities that would otherwise obstruct the amplification process. The leaf, commonly used for nucleic acid extraction has layers of cuticle and cell wall covering it. The extraction process becomes difficult by the requirement to breakdown these layers to remove the genetic material within. Also, a variety of contaminants such as proteins, polysaccharides, polyphenolics, and several secondary metabolites may be present. It is essential to eliminate these impurities from the genetic material in order to produce a high-quality template that can be amplified. These cellular proteins are removed by organic solvents like phenol and chloroform, however integrating such steps in portable device is difficult [14]. In some cases, plant crude extract combined with extraction buffer is directly used for amplification. detected Phytophthora castoreum in strawberries using crude crown tissue macerated with ELISA grinding buffer coupled with RPA. Conducted an experiment where they used crude extract and pure bacterial DNA template for detection of Xanthomonas fragariae [15]. Successful LAMP DNA amplification was achieved using both crude extract and pure bacterial DNA control. Thus, utilization of crude extract for extraction protocol is simple but potentially lacks sensitivity. So, sensitivity and specificity of protocol can be enhanced by purifying target DNA. Another rapid and efficient DNA extraction method is LFD that has been successfully used for POC testing. Here, sample is disintegrated with the help of metal ball bearings in extraction buffer and then transferred on to the pad of nitrocellulose membrane. After that a tiny portion of nitrocellulose membrane is cut-off and placed to the reaction mix for amplification [16]. In this case, isolated DNA remains stable on membrane at room temperature, that makes it acceptable for use in field [17]. This method has been used for detection of Phytophthora ramorum and Phytophthora kernoviae.
Another DNA extraction technique with possible POC use is solid phase reversible immobilization (SPRI). It requires only magnet and a micropipette. After macerating the sample in lysis buffer using a plastic pestle, the lysate was filtered through a pipette tip to eliminate any remaining cell debris [18]. DNA is then purified from the lysate using SPRI and the extracted DNA was utilized directly for amplification. This technique has numerous applications in detecting human and plant diseases. In this technique, it becomes possible to extract DNA without centrifugation steps, it just requires a magnet that is portable and can be utilized on site, even in places lacking power supply [19]. Another exceedingly important extraction method having POC use is cellulose-based dipstick where sample processing time has been reduced to 30s. Plant tissues are macerated for 8-10s by shaking in a tube with extraction buffer and one or two ball bearings. After that a cellulose dipstick is inserted in the sample tube followed by rinsing three times in wash buffer and finally placed into the tube containing the amplification reaction mixture. This method can be easily coupled with many amplification techniques such as PCR, LAMP, and RPA. This technique is just as sensitive as SPRI but is less expensive, rapid and eliminates the need for pipetting steps [20]. Recently, there has been a rise in new methods for extracting nucleic acids for POC analysis. investigated a variety of materials to determine their ability to trap nucleic acids [21-25]. They observed that Whatman No. 1 cellulose-based filter paper effectively entraps and retains DNA without the requirement of any chemical treatment. These paper-based extraction methods were successfully evaluated on different plant species, such as wheat, rice, tomato, soybean, tobacco, mandarin, and lemon, demonstrating good sensitivity. These membrane-based extraction technologies such, Flinders Technology Associates cards (FTA cards), and alumina membranes escape solvents and directly proceed to amplification of nucleic acids bound to the membrane. Paper-based microfluidics device has become a viable multiplexable POC platform, potentially valuable in resource limited settings. This method has found application in food safety, veterinary medicine, health care, and environmental and crop monitoring. It is based on direct virus absorption onto PCR tubes at 4◦C. This extraction procedure provides viral RNA suitable for molecular analysis within 5 min. Although this procedure is relatively quick and easy to use, more research is still required to see whether it could be used for routine application to different pathogens [26-30].
Thermal Cycling Amplification
PCR, along with its variants quantitative PCR, reverse transcription-PCR, nested PCR, and digital PCR. etc. are undoubtedly well known and extensively studied techniques for amplifying nucleic acids. It targets particular sequences in genomic DNA to detect and identify plant pathogens. Although, PCR is very reliable, sensitive, robust and rapid but it is limited by several technical issues when basic POC amplification is needed. To accomplish thermal cycling at three different temperatures for strand separation, primer annealing and extension of target sequence, it demands a thermal cycler, a device that is difficult to miniaturize. This thermal cycling is energy requiring process that is not provide by battery power supply which ultimately limits its application as POC diagnostics. Still some fascinating attempts have been developed for its POC application. For example, Schwenkbier used linearafter- the-exponential PCR (LATE-PCR) as amplification technique for on-site detection of Phytophthora species [31]. Developed a protocol for on-site detection of Xylella fastidiosa using a portable Smart Cycler. Similarly, the protocol for on-site detection of pathogens using qPCR have been described for Plum pox virus Spongospora subterranean, Austropuccinia psidii and Pratylenchus penetrans. Recently, iSPEED (In Situ Processing and Efficient Environmental Detection) kit have been developed to detect pathogens like Sphaerulina musiva, Cronartium ribicola, C. comandrae, Phytophthora lateralis, and P. ramorum. of forest trees using POC qPCR. This kit is compact enough be carried in a small backpack and is an efficient solution for in field diagnostics. Many isothermal techniques have been emerged as alternatives to thermal cycler although each one has advantages and disadvantages. These methods operate at a single temperature that obviate the need for complex heating and cooling steps. Even though some of the other technologies are isothermal in nature, they are not appropriate for POC applications since they are practically complex and require many pipetting procedures [32]. The four most well-known and effective isothermal amplification techniques having POC applications include LAMP, RPA, HDA, and RCA.
Loop-mediated isothermal amplification
Loop-mediated isothermal amplification (LAMP) is a nucleic acid amplification method pioneered by Notomi, which has been extensively used for many medical and agricultural domains. In case of LAMP, DNA synthesis is best accomplished at a temperature range between 60 and 65°C using Bst polymerase, a DNA polymerase derived from Bacillus stearothermophilus. During the amplification process, LAMP uses a minimum of four primers (forward inner primer, forward outer primer, backward inner primer, backward outer primer) to identify six distinct sequences in the target DNA. The forward inner primer containing sense and antisense sequences in the DNA, hybridizes the target sequence and initiates DNA synthesis. Next, the forward outer primer facilitates the stranddisplacement and creates a single stranded DNA molecule that functions as a template for backward inner primer and backward outer primer and finally producing a ssDNA molecule with a loop structure. LAMP offers several advantages that makes it suitable for POC applications. It was able to detect plum pox virus in with in short time of 2.5 h at constant temperature and is relatively cost effective. Moreover, reagents used in LAMP are thermally stable at nonoptimal storage temperatures maintaining their efficiency even when stored at 25°C or 37°C for 15 days compared to controls stored at −20°C. The amplification efficiency is exceptionally robust, sensitive and specific yielding a huge amount of PCR product with minimal amount of input DNA. While LAMP offers substantial advantage for onsite detection of pathogen, it also has some notable drawbacks. According to LAMP primer designing is complicated and the use of nonoptimal primers can result in a number of technical problems, including inadequate template amplification (false negatives) and the production of nonspecific amplicons (false positives). Despite the availability of software for primer designing, it is not assured that primers will perform optimally and also many published primers sets for particular pathogens do not perform efficiently when replicated in laboratory settings, because of variations in the nucleic acid extraction procedure or some other unidentified and uncontrollable factors. In LAMP procedure, the use of DMSO and betaine reduces nonspecific amplification [33]. LAMP-based diagnostics have been developed for rice to identify viral, bacterial and fungal diseases such as Rice stripe virus, and Rice black-streaked dwarf virus, bacterial blight (Xanthomonas oryzae pv. oryzae), bacterial leaf streak (Xanthomonas oryzae pv. oryzicola), rice blast (Magnaporthe oryzae), sheath blight (Rhizoctonia solani). Also, several other pathogens like Puccinia triticina, Pyricularia oryzae, and Fusarium graminearum, are also detected using LAMP- based approaches. LAMP based methods safeguard national boarders from introduction of new pest and pathogen, offers rapid POC systems for quarantine and inspection services. Many fast LAMP protocols for POC detection of plant pathogens have been developed. All these protocols unite fast nucleic acid extraction techniques and signal readout to enable LAMP to carry out all the required steps on site.
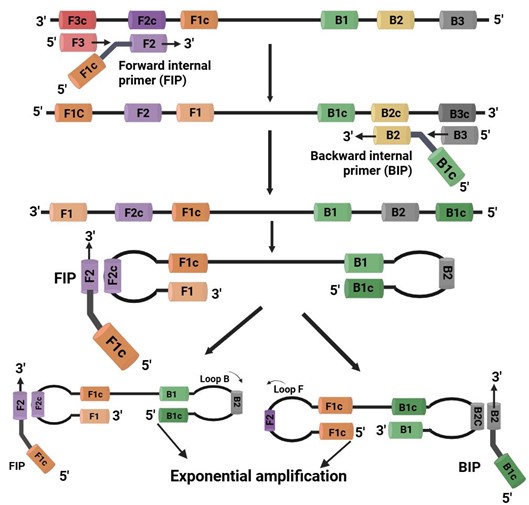
Figure 1: Schematic outline of loop-mediated isothermal amplification (LAMP). LAMP uses two sets of primers targeting six distinct regions of the DNA. The inner primers, containing both sense and antisense sequences, initiate DNA synthesis by hybridizing the target. The outer primers then perform stranddisplacement synthesis, generating single-stranded DNA templates for the second set of primers. This forms a looped DNA structure. The inner primers bind to the loop, initiating synthesis of new strand [34].
Recombinase Polymerase Amplification
Recombinase polymerase amplification (RPA) is another isothermal technique that involves enzymatic mechanism to unwind double-stranded DNA (dsDNA) to facilitate primer binding to the target DNA, avoiding the need for an initial heating step to unwind the target DNA. Unlike PCR, unwinding of whole dsDNA template is not necessary for primer binding as unwinding of short complementary target site is enough. The reaction starts by integrating recombinase protein with forward and reverse primers that assists them in locating the correct target and unwinds a part of doublestranded DNA. After primer binding, the structure formed is balanced by the binding of a single-strand binding (SSB) protein to prevent primer ejection. Once the recombinase gets dissociated from primers, making their 3′ ends accessible, that enables the Bacillus subtilis Pol I DNA polymerase to start transcription which is also known have a strand displacement activity. The RPA technology generates millions of DNA copies within short reaction time and at a constant temperature of 37◦C and 42◦C. This low temperature is supplied by simple battery powered devices, or even in some studies, successful nucleic acid amplification has been reported using human body heat [35]. In comparison to LAMP, the primer design in RPA is simpler and allows for multiplexing to identify many pathogens. This technology is extremely sensitive, with low detection limit of 6.25 fg of input DNA and specificity exceeding 95%. The reagents used during reaction process are thermally stable and even after 12 weeks at 25°C, they did not exhibit a drop in amplification efficiency. All these advantages make this technique an ideal for POC in remote locations. However, RPA have some limitations, such as fluctuating sensitivity and its ability to amplify only small DNA fragments (500bp) using, which leads to non-specific amplification. By integrating RPA with CRISPR-based technologies like SHERLOCK, DETECTR, and iSCAN, its sensitivity can be enhanced. RPA coupled with surface-enhanced Raman scattering and electrochemical biosensors emerged as a new technology to detect pathogens like Pseudomonas syringae, Botrytis cinerea, and F. oxysporum in plants. In order to identify RNA viruses (maize chlorotic mottle virus) and viroids (tomato apical stunt viroid), RPA was coupled with initial initial RT step coupled RT-RPA and lateral flow to detect cherry virus from crude extracts, that proved to be more economical than RT-PCR [36]. Commercial RPA-based diagnostic kits are available for many plant pathogens such as Banana bunchy top virus and F. oxysporum f. sp. vasinfectum, Race 4. A low-cost prototype known as the POCKET (Point-Of-Care Kit for the Entire Test) was recently developed that integrated RPA, microfluidics, 3D printing, and a smartphone technology, to form a portable device for assessing various DNA samples. All the abovementioned RPA-based techniques can be tailored for POC use by combining with an effective nucleic acid extraction technique and a suitable POC readout.
Helicase-Dependent Amplification In 2004, New England Biolabs developed an isothermal approach called as Helicase dependent amplification (HAD). Its principle is very simple and same as that of conventional PCR but it does not need heating step to denature the dsDNA. Instead, DNA is denatured with the help of enzyme helicase letting forward and reverse primers to bind at target site to start synthesis at isothermal temperature (37°C). In order to stop rehybridization of complementary ssDNA molecules, SSB protein and MutL endonuclease are added to the reaction and quantifiable amount of PCR amplicons are produced within 60 min. HDA's features make it perfect for POC use, but it also has some significant drawbacks, such as the need for complex optimization to ensure coordinated enzyme activity between the helicase and DNA polymerase and the possibility of nonspecific amplification due to template-independent primer interactions. By using betaine and DMSO in the reaction, the nonspecific amplification can be decreased, but the sensitivity is lost in the process. Furthermore, the final outcome may be strongly impacted by the presence of SSB and MutL, which are necessary to stop ssDNA from re-hybridizing to create dsDNA [37]. Many studies have reported that HDA is ineffective to amplify lengthy targets that is attributed to restricted unwinding speed (20 bp/s) of UvrD and process less than 100 bp per binding Also, MutL does not improve the processing rate, despite increasing UvrD unwinding activity. Recently HDA diagnostics have made advancement after discovering thermostable UvrD helicase (Tte- UvrD) from Thermoanaerobacter tengcongensis that amplifies target DNA at higher temperature, finally decreasing rehybridization of ssDNA and thereby eliminating the requirement of SSB and MutL [38]. HDA -based diagnostics have clinical application in detecting pathogens like Salmonella paratyphi and diarrhea-causing pathogens, as well as have veterinary applications such as identifying Streptococcus equi causing strangles in horses. However, its use in plant pathology is rare and is limited to the identification of citrus leprosis virus C and tobacco mosaic virus. To enhance the sensitivity of HDA, it has been coupled with other techniques like ELISA (Gill et al., 2007) and gold nanoparticles to detect Helicobacter pylori. The specificity and sensitivity showed 90% increase in both cases in comparison to original HDA assay.
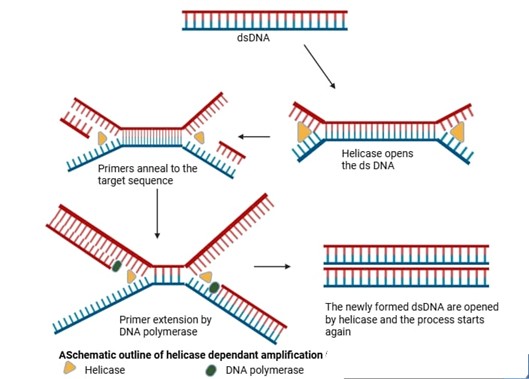
Figure 2: Schematic Representation of Helicase-Dependent Amplification (HDA)
Rolling Circle Amplification
RCA is a pioneer isothermal process that is used to amplify circular DNA. A single primer is annealed to a circular DNA template and is extended by the strand displacement activity of ϕ29 DNA polymerase, which causes newly synthesized DNA to displace previously generated DNA and releases ssDNA. This freshly formed long ssDNA contains repeated sequence complementary to the circular template. In RCA, linear DNA is also used as a template for reaction. Firstly, padlock probes are designed in such a that hybridize with the target sequence to take on a padlock- like circular conformation. The circular ssDNA molecule is then formed by ligating the probe molecule with T4 ligase. After that primer binds to this newly synthesized ssDNA molecule. The padlock probes have high multiplexing efficiency, so used for detection of many plant pathogens at the same time. This technique is highly specific and less prone to non-specific amplification as no new 3’ -end ssDNA is produced which could otherwise act as a primer for non-specific amplification [39]. RCA is coupled with other techniques like RFLP and direct sequencing for identifying and classifying phytopathogens at cheaper cost. Used visual inspection of RCA products to identify over 40 strains of Fusarium spp. Additionally, compared to gel electrophoresis, the sensitivity has been greatly increased by combining RCA with other readout technologies as immunological tests, DNA biosensors, and microarrays. This method is too labor-intensive for POC applications because it requires numerous hybridization and enzymatic stages for the synthesis of padlock and RCA amplification.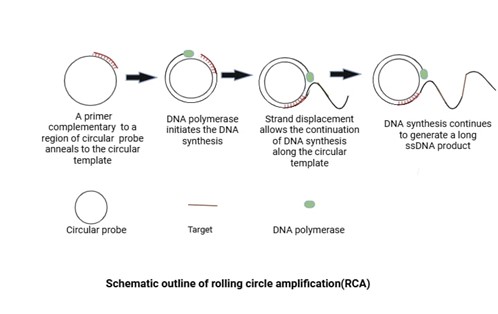
Figure 3: Schematic Representation of Rolling Circle Amplification (RCA)
Table 1: List of methods applied for POC plant pathogen detection.
| Method | Advantages | Disadvantages | Pathogen detected | References |
|---|---|---|---|---|
| Loop-mediated isothermal amplification | Rapid, isothermal, high sensitivity, relatively cost effective | Complexity of Primer design | Phytophthora ramorum, P. kernoviae, Phytophthora infestans | Tomlinson et al. (2010); Ammour et al. (2017); Aglietti et al. (2019) |
| Recombinase polymerase amplification | Rapid, isothermal, and eliminates the need for an initial denaturation step. | Requires Long primers that results in variability in specificity and sensitivity | Verticillium alfalfae, Fusarium oxysporum f. sp. fragariae | Gaige et al. (2018); Burkhardt et al. (2019); Strayer-Scherer et al. (2019) |
| Rolling circle amplification | Isothermal, highly sensitive and specific | Costly with tricky detection | Watermelon chlorotic stunt virus, Fusarium graminearum | Davari et al. (2012); Rezk et al. (2019) |
| Helicase-dependent amplification | Fast, isothermal, does not need an initial denaturation step | Demands high optimization | Phytophthora kernoviae, Tomato spotted wilt virus | Schwenkbier et al. (2015); Wu et al. (2016) |
| Fluorescence based detection | Reliable, sensitive, easy to incorporate into microfluidic devices | Expensive, not visible by naked eye | Banna viruses, Xanthomonas euvesicatoria | Larrea-Sarmiento et al. (2018); Lei et al. (2019); Xia et al. (2019); Zhang et al. (2019) |
| Colorimetric detection | Cheap, minimal equipment requirement. | Reduced sensitivity and specificity | Alternaria panax | Kolm et al. (2015); Lin et al. (2015); Wei et al. (2018) |
| MinION sequencing | High-throughput results, efficient detection of multiple pathogens | Not completely user friendly | Phytophthora capsici | Liau et al. (2019); Shaffer (2019); Cui et al. (2019) |
POC AMPLIFICATION READOUTS
Amplicons can be detected via fluorescence, chemiluminescence, and colorimetric visualization. An amplification readout device can expedite detection analysis in the field for point-of-care (POC) diagnostic procedures by requiring minimal instruments. The majority of these devices use optical or visual tools, such as light detectors, smartphones, and CCD cameras, so the signal may be seen with the unaided eye. SYBR Green I, EvaGreen, and a family of cyanine SYTO dyes that bind to dsDNA are examples of intercalating dyes that must be used with fluorescence-based detection techniques. They are helpful for amplicon detection in a timely manner because the binding boosts fluorescence intensity. Commonly used dyes in LAMP reactions are EvaGreen and SYBR Green I, however high reagent concentrations significantly impede the reaction process. Fluorescence detectors and amplification techniques, such as qPCR, LAMP, have been combined in a multitude of portable devices. Surface-enhanced Raman scattering (SERS) for fast diagnostic assays is another potent and extremely promising readout method. With just one laser excitation, this method yields improved Raman scattering patterns of the molecules adsorbed from metal nanoparticle surfaces. For highly multiplexed applications, SERS yields narrow and distinct spectral peaks, which, when analyzed, provide more precision than typical fluorescent-based approaches. In order to identify several plant diseases infecting Arabidopsis thaliana at an early stage of infection, multiplex RPA was combined with SERS in 2016. With this method, multiplex field detection can be completed in 90 minutes at a single, continuous low temperature of 37°C with excellent sensitivity and specificity. However, the implementation of this technology is limited by the high cost of portable SERS readers [40]. LAMP amplification results in an insoluble magnesium pyrophosphate, which raises the turbidity of the reaction mixture in colorimetric readout techniques. The spectrophotometric measurement of light scattering brought on by turbidity has been employed as an accurate indicator of amplification, and the rise in turbidity is proportionate to the amount of DNA produced. However, turbidity can only be seen at extreme amplifications and requires an expert operator to notice with the unaided eye. A considerable amount of insoluble magnesium pyrophosphate is produced by LAMP reactions in colorimetric readout systems. This insoluble magnesium pyrophosphate stays suspended in the amplification reaction mixture, increasing turbidity. A reliable indicator of amplification is the spectrophotometric measurement of light scattered by the turbid reaction mix, which increases in proportion to the amount of DNA produced. However, it still has some drawbacks, mainly lack of sensitivity and specificity. Lateral flow assays (LFAs) can sometimes be seen as amplification readouts, and because they provide a simple yes/no readout, they are appropriate for point-of-care (POC) applications. Originally intended to be used with particular antibodies to identify proteins and peptides, LFAs have now developed to identify a wide range of other biological components, including DNA. In plant diagnostics, LFAs in conjunction with isothermal amplification have been frequently utilized. RPA-LFA techniques have been developed for the identification of bacterial, viral, and phytoplasma diseases in a variety of crops, including citrus, pepper, potato, soy, and strawberry. There are other multiplex LAMP-LFA techniques available, such as the simultaneous detection of Potato virus X and Tobacco rattle virus in potatoes.
POC SEQUENCING
Without any prior understanding of the pathogens' nature, nextgeneration sequencing (NGS) has the enormous potential to discover many pathogens in a single analysis procedure. NGS has been widely employed in the field of plant pathology, particularly for virus identification, to identify viruses without culturing and by directly utilizing ambient materials. Nonetheless, most NGS tests are conducted in specialist labs and necessitate costly apparatus, a dependable power source, sample preparation, and intricate data processing algorithms. Therefore, POC routine diagnostic analyses cannot afford the present cost of NGS sequencing technologies; nevertheless, in recent years, portable sequencers have been created that enable low-cost amplicon sequencing to be performed directly in the field. Nanopore sequencing, sometimes referred to as fourthgeneration DNA sequencing technology, is one such instance. It works by electrophoretically moving electrolyte ions in a solution from one side of a nanopore to the other via the introduction of an external voltage. The ion current is stopped when DNA or RNA molecules are introduced to the solution because they are forced through nanopores by an external voltage. Real-time molecular sequencing is made possible by changes in ionic current during translocation, which correlate to nucleotide sequences that flow through the pore and are deciphered by computer algorithms. Oxford Nanopore Technologies tested MinION, a real-time portable DNA and RNA sequencing device, in 2014. It uses a high-speed USB connector to connect to a PC or laptop and weighs less than 100 g. With the help of replaceable flow cells that immobilize the nanopores, this apparatus can quantify the speed at which a DNA strand passes through biological nanopores and identify DNA molecule translocations with single-nucleotide accuracy. The MinION uses Metrichor, an online cloud-based analysis platform, to do base-calling by analyzing sequences that are read by the MinION in real-time. Workflows such as "What's in my pot" (WIMP) have been developed for species identification. WIMP uses a real-time species identification bioinformatics application in conjunction with MinION technology to identify and classify bacteria in about 3.5 hours. The open-source base caller Nanocall was created in 2016 and offers users an offline analysis source for data provided by MinION, making it appropriate for usage in remote areas. In 2020, Boza et al. developed the DeepNano-blitz base caller, which is capable of performing real-time data processing in the field without the need for a strong IT facility. Additional instances of MinION's application in agriculture and forestry comprise the identification of pathogenic fungi, bacteria, viruses, and nematodes [41]. Even in scenarios when resources are scarce, the MinION device has been utilized in conjunction with a few technologies for identification and detection. For instance, Imai et al. (2017) identified malaria in blood samples utilizing LAMP amplicon sequencing with the MinION and FTA cards for DNA extraction. Point-of-care sequencing is a rapidly developing technology that has demonstrated rapid advancements in mobility, accuracy of sequencing, and ease of use. Because of all these qualities, point-of-care sequencing (POC sequencing) may prove to be a viable substitute for plant pathogen detection in the future.
CONCLUSION AND FUTURE PERSPECTIVES
POC diagnostic testing's primary advantage is its ability to deliver fast results on-site, allowing farmers to avoid crop losses and make quick management decisions. POC diagnostic kits must be easy to carry and use so that even a single person without a background in science may perform the test. Numerous studies have demonstrated that, despite the great majority of current applications being PCR-based, existing isothermal approaches can perform on par with or even better than PCR-based tests. Isothermal techniques are attractive options for point-of-care diagnostic assays in low resource environments as they can perform reactions at a constant temperature. The ongoing development of portable and sensitive nucleic acid detection methods is a rapidly expanding field, and will provide an exciting possibility for both researchers and farmers in the coming years. POC analysis frequently offers quick disease diagnosis, low risk, cheap cost, and an excellent diagnostic experience for the operator. However, several challenges remain, including developing reliable methods for all major pathogens that pose a global threat to agriculture, tailoring these methods to the specific needs of different plants and pathogens, providing a strong comparison with conventional technologies, and persuading end users to switch from conventional detection methods to innovative ones that offer substantial advantages.
